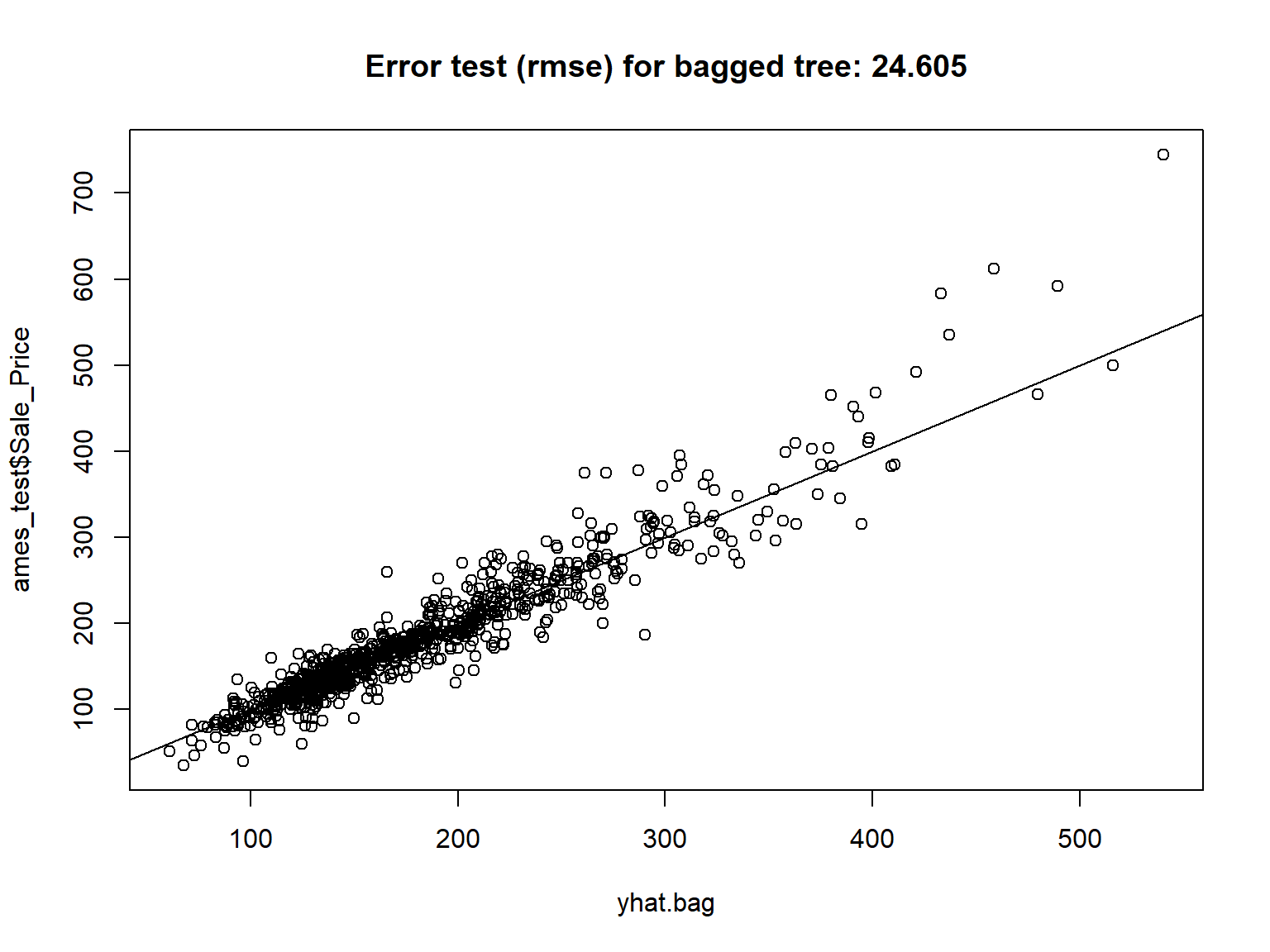
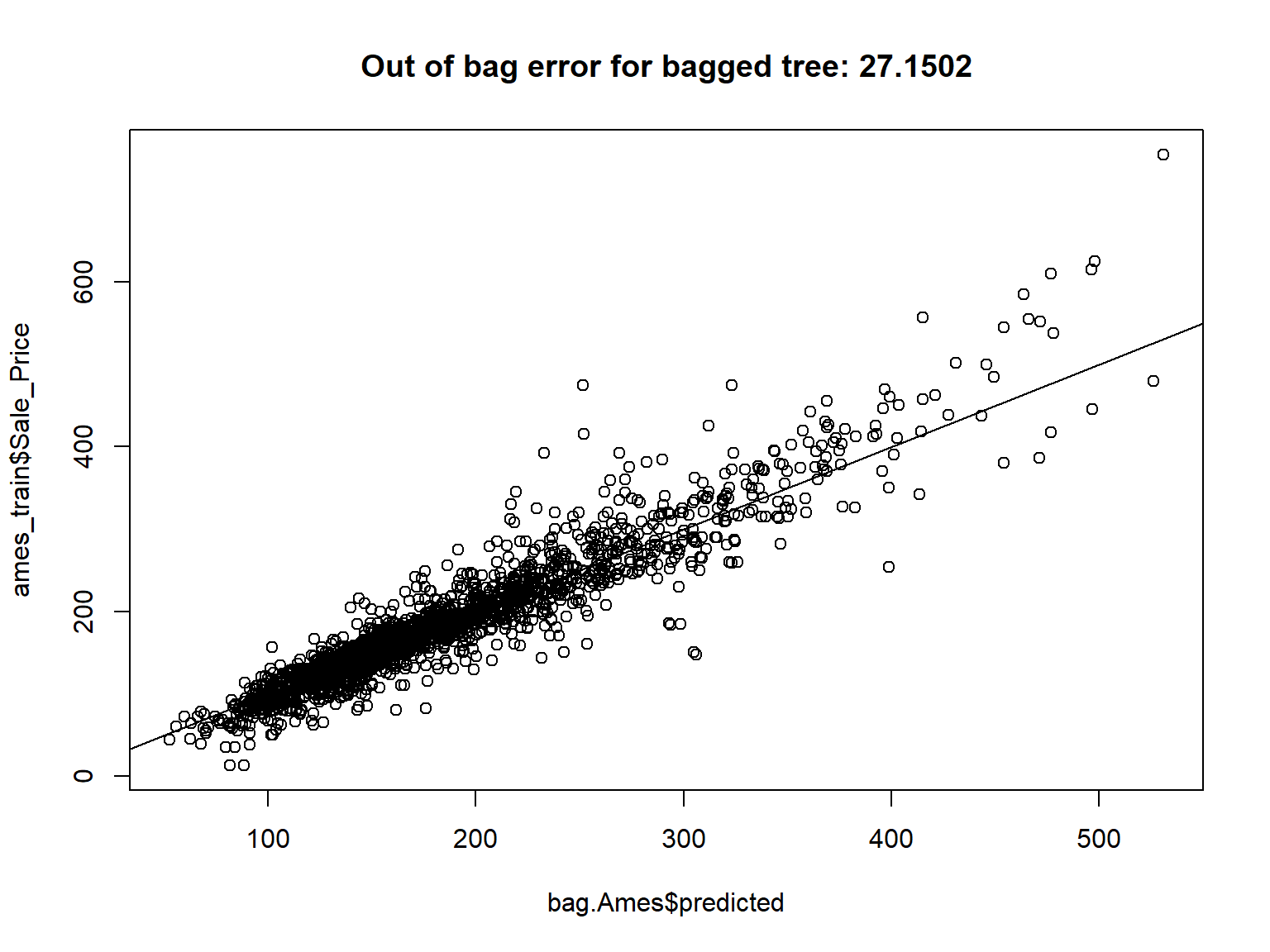
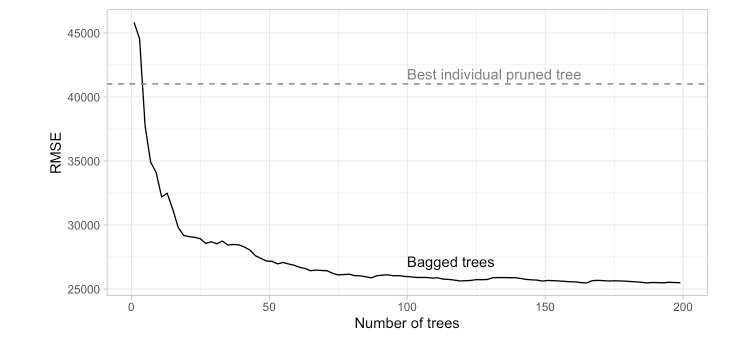
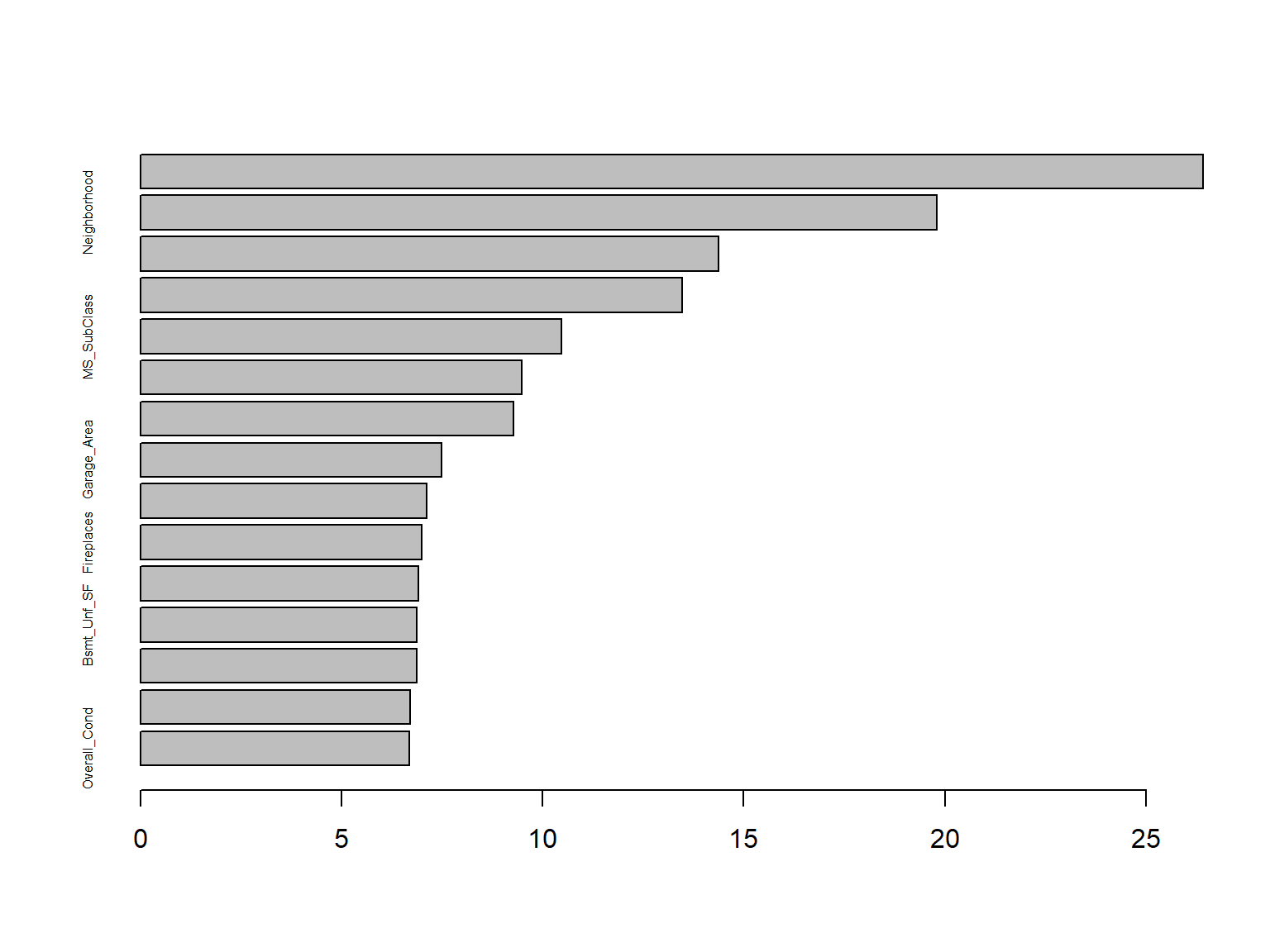
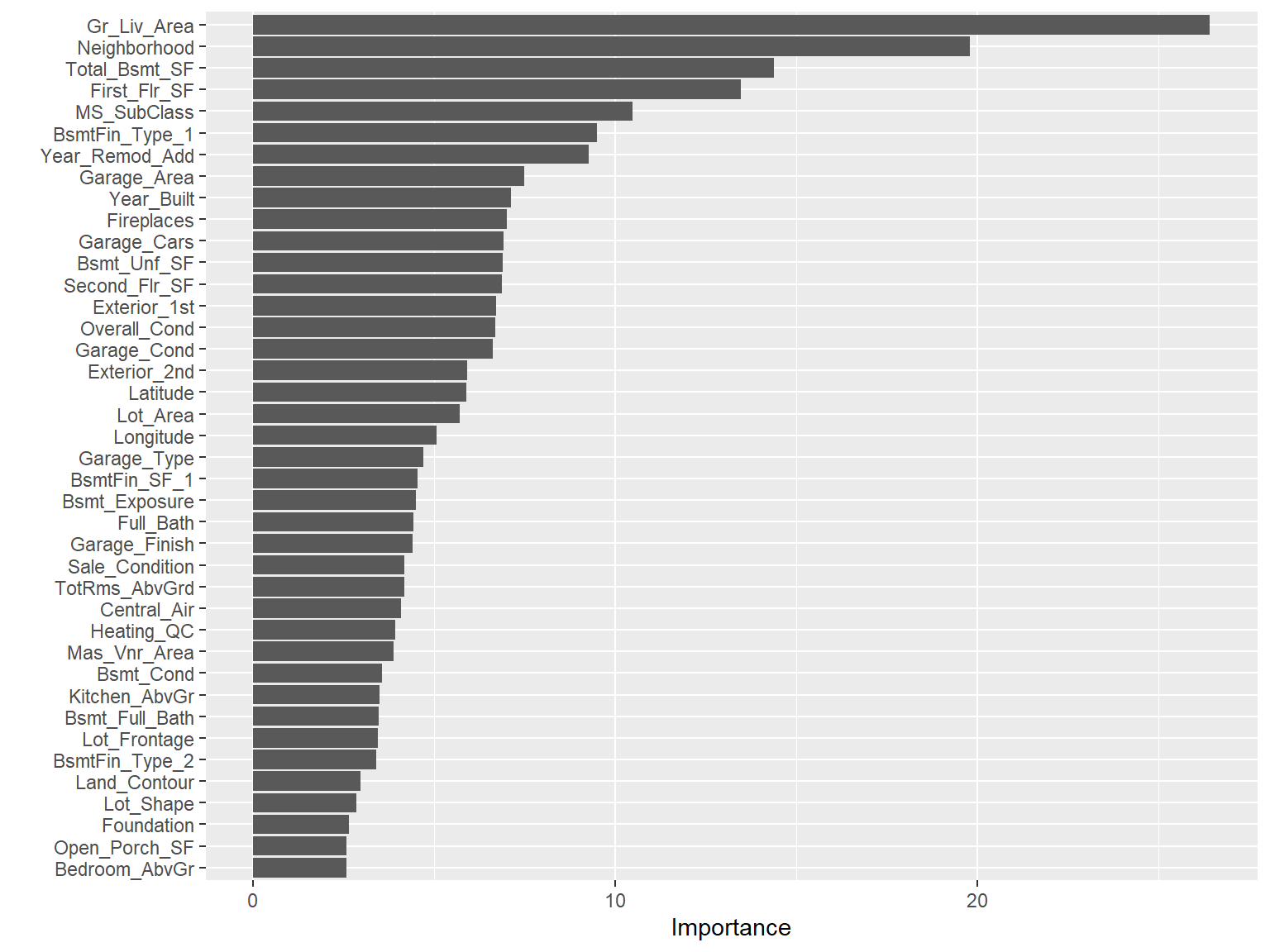
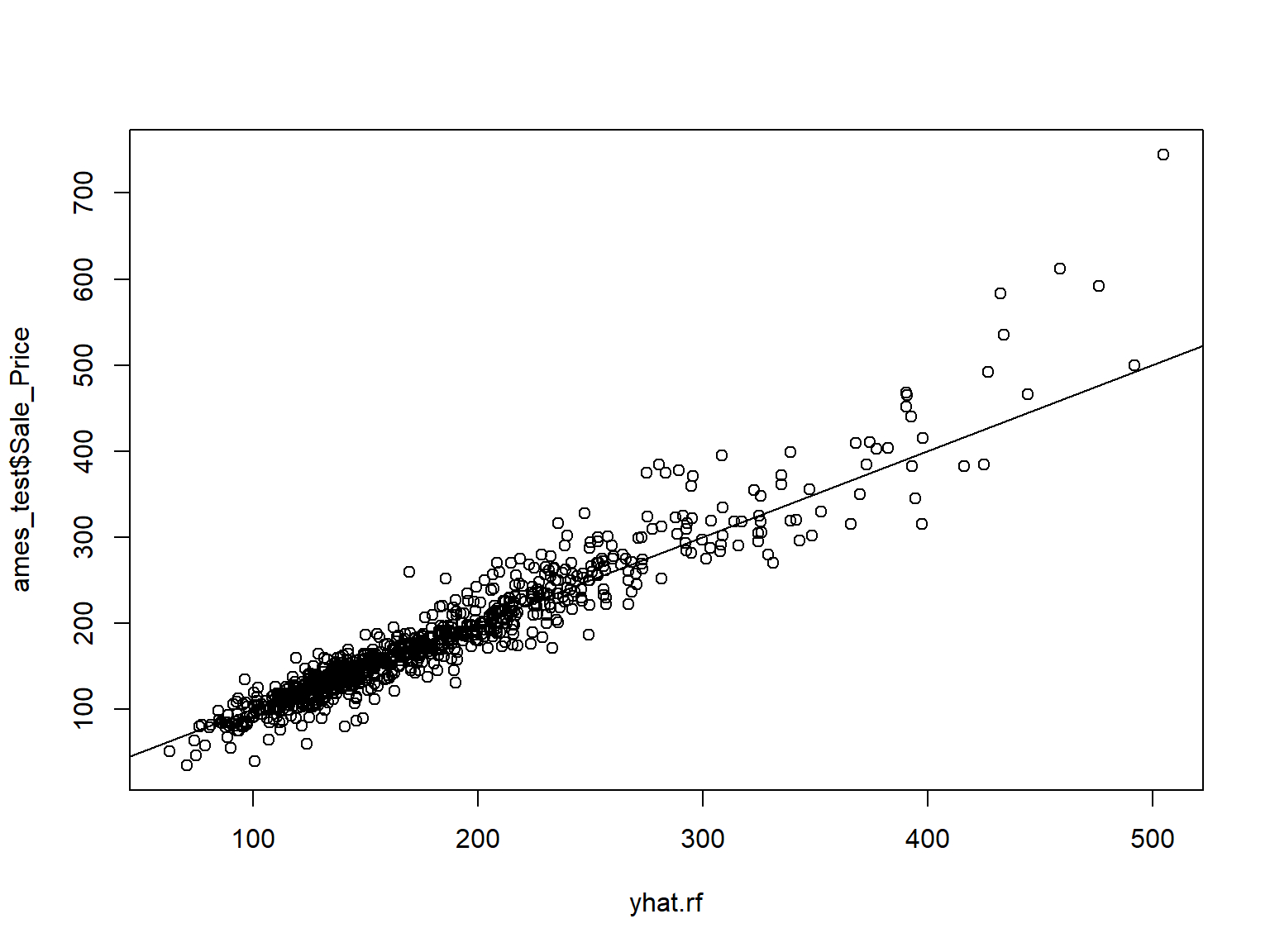
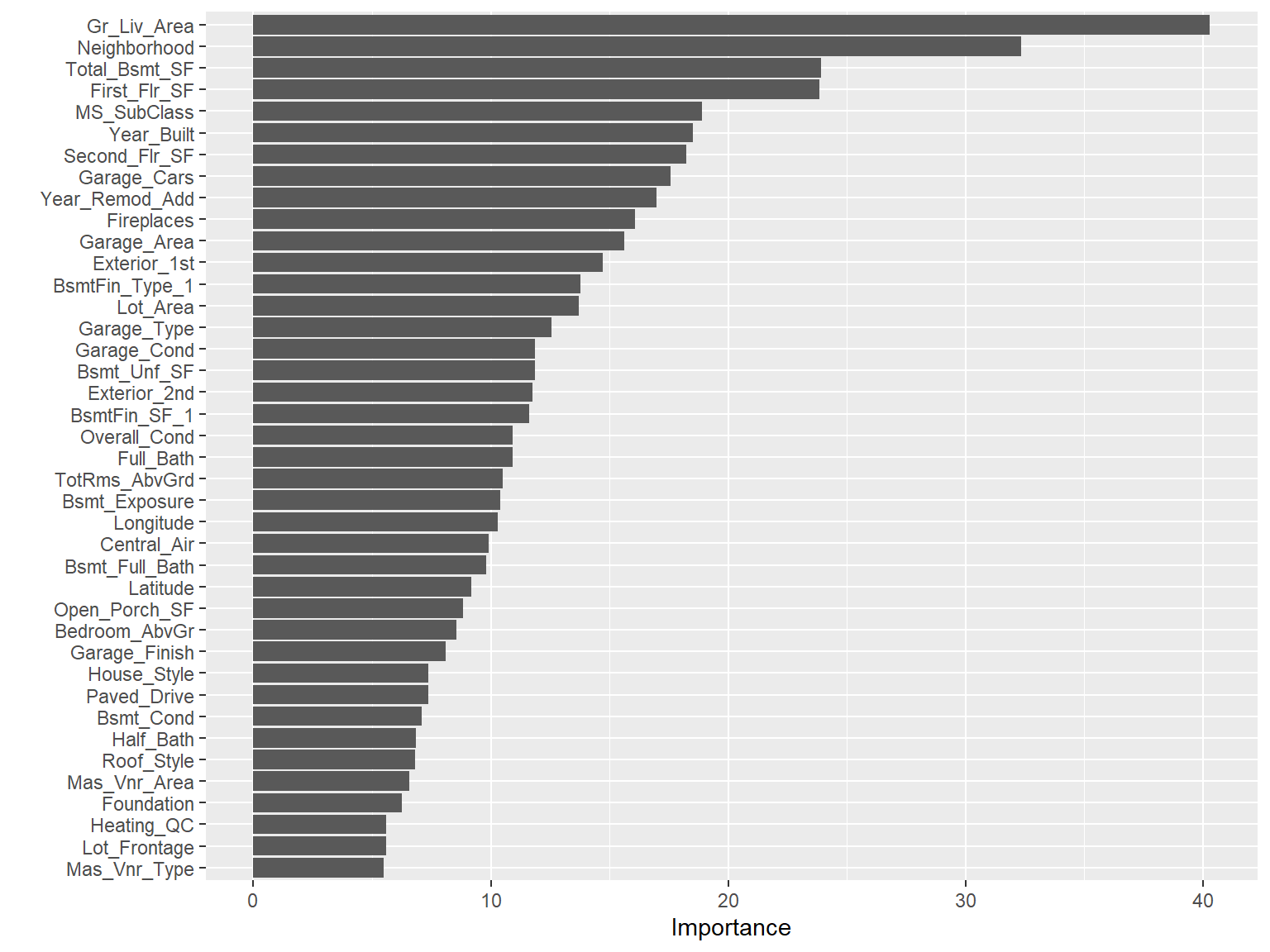
| MS_SubClass |
0 |
1 |
FALSE |
16 |
One: 1079, Two: 575, One: 287, One: 192 |
| MS_Zoning |
0 |
1 |
FALSE |
7 |
Res: 2273, Res: 462, Flo: 139, Res: 27 |
| Street |
0 |
1 |
FALSE |
2 |
Pav: 2918, Grv: 12 |
| Alley |
0 |
1 |
FALSE |
3 |
No_: 2732, Gra: 120, Pav: 78 |
| Lot_Shape |
0 |
1 |
FALSE |
4 |
Reg: 1859, Sli: 979, Mod: 76, Irr: 16 |
| Land_Contour |
0 |
1 |
FALSE |
4 |
Lvl: 2633, HLS: 120, Bnk: 117, Low: 60 |
| Utilities |
0 |
1 |
FALSE |
3 |
All: 2927, NoS: 2, NoS: 1 |
| Lot_Config |
0 |
1 |
FALSE |
5 |
Ins: 2140, Cor: 511, Cul: 180, FR2: 85 |
| Land_Slope |
0 |
1 |
FALSE |
3 |
Gtl: 2789, Mod: 125, Sev: 16 |
| Neighborhood |
0 |
1 |
FALSE |
28 |
Nor: 443, Col: 267, Old: 239, Edw: 194 |
| Condition_1 |
0 |
1 |
FALSE |
9 |
Nor: 2522, Fee: 164, Art: 92, RRA: 50 |
| Condition_2 |
0 |
1 |
FALSE |
8 |
Nor: 2900, Fee: 13, Art: 5, Pos: 4 |
| Bldg_Type |
0 |
1 |
FALSE |
5 |
One: 2425, Twn: 233, Dup: 109, Twn: 101 |
| House_Style |
0 |
1 |
FALSE |
8 |
One: 1481, Two: 873, One: 314, SLv: 128 |
| Overall_Cond |
0 |
1 |
FALSE |
9 |
Ave: 1654, Abo: 533, Goo: 390, Ver: 144 |
| Roof_Style |
0 |
1 |
FALSE |
6 |
Gab: 2321, Hip: 551, Gam: 22, Fla: 20 |
| Roof_Matl |
0 |
1 |
FALSE |
8 |
Com: 2887, Tar: 23, WdS: 9, WdS: 7 |
| Exterior_1st |
0 |
1 |
FALSE |
16 |
Vin: 1026, Met: 450, HdB: 442, Wd : 420 |
| Exterior_2nd |
0 |
1 |
FALSE |
17 |
Vin: 1015, Met: 447, HdB: 406, Wd : 397 |
| Mas_Vnr_Type |
0 |
1 |
FALSE |
5 |
Non: 1775, Brk: 880, Sto: 249, Brk: 25 |
| Exter_Cond |
0 |
1 |
FALSE |
5 |
Typ: 2549, Goo: 299, Fai: 67, Exc: 12 |
| Foundation |
0 |
1 |
FALSE |
6 |
PCo: 1310, CBl: 1244, Brk: 311, Sla: 49 |
| Bsmt_Cond |
0 |
1 |
FALSE |
6 |
Typ: 2616, Goo: 122, Fai: 104, No_: 80 |
| Bsmt_Exposure |
0 |
1 |
FALSE |
5 |
No: 1906, Av: 418, Gd: 284, Mn: 239 |
| BsmtFin_Type_1 |
0 |
1 |
FALSE |
7 |
GLQ: 859, Unf: 851, ALQ: 429, Rec: 288 |
| BsmtFin_Type_2 |
0 |
1 |
FALSE |
7 |
Unf: 2499, Rec: 106, LwQ: 89, No_: 81 |
| Heating |
0 |
1 |
FALSE |
6 |
Gas: 2885, Gas: 27, Gra: 9, Wal: 6 |
| Heating_QC |
0 |
1 |
FALSE |
5 |
Exc: 1495, Typ: 864, Goo: 476, Fai: 92 |
| Central_Air |
0 |
1 |
FALSE |
2 |
Y: 2734, N: 196 |
| Electrical |
0 |
1 |
FALSE |
6 |
SBr: 2682, Fus: 188, Fus: 50, Fus: 8 |
| Functional |
0 |
1 |
FALSE |
8 |
Typ: 2728, Min: 70, Min: 65, Mod: 35 |
| Garage_Type |
0 |
1 |
FALSE |
7 |
Att: 1731, Det: 782, Bui: 186, No_: 157 |
| Garage_Finish |
0 |
1 |
FALSE |
4 |
Unf: 1231, RFn: 812, Fin: 728, No_: 159 |
| Garage_Cond |
0 |
1 |
FALSE |
6 |
Typ: 2665, No_: 159, Fai: 74, Goo: 15 |
| Paved_Drive |
0 |
1 |
FALSE |
3 |
Pav: 2652, Dir: 216, Par: 62 |
| Pool_QC |
0 |
1 |
FALSE |
5 |
No_: 2917, Exc: 4, Goo: 4, Typ: 3 |
| Fence |
0 |
1 |
FALSE |
5 |
No_: 2358, Min: 330, Goo: 118, Goo: 112 |
| Misc_Feature |
0 |
1 |
FALSE |
6 |
Non: 2824, She: 95, Gar: 5, Oth: 4 |
| Sale_Type |
0 |
1 |
FALSE |
10 |
WD : 2536, New: 239, COD: 87, Con: 26 |
| Sale_Condition |
0 |
1 |
FALSE |
6 |
Nor: 2413, Par: 245, Abn: 190, Fam: 46 |